PROJECT #1 INDEXING SUBSTRINGS
By Pamela Cutter • Edited by Sandino Vargas-Pérez
In this project, you will write a program that will report each n-letter substring that appears in a list of strings, as well as where it appears in each of the strings, where n is to be determined by the user. This is a process that is useful in finding shared substrings in DNA sequences.
For example, suppose we have a list of strings: acgtacgtacgtacgt, cgtacgtacgtacgt, gtacgtacgtacgtacgtacgt, and tacgtacgtacgt, and we are looking for 4-letter substrings. The first substring we find, acgt, appears:
- 4 times in the first string at positions 0, 4, 8, and 12.
- 3 times in the second string at positions 3, 7, and 11.
- 5 times in the third string at positions 2, 6, 10, 14, and 18.
- 3 times in the fourth string at positions 1, 5, and 9.
acgt. It will then report the 12 occurrences of cgta, the next substring encountered, and will continue with the occurrences of gtac, etc.
GETTING STARTED
There are a number of files provided to get you started. To begin, download the following files and create a project in BlueJ (or Eclipse) containing them.
- DNAData.txt: This is a file of partial DNA sequences, taken from a set of viruses. A DNA sequence consists of a GI identifier, a description, and then a sequence of nucleotides. These nucleotide sequences are the strings that you will be indexing.
-
TestData.txt: This file has a format similar to
DNAData.txt, but can be used to easily check the correctness of your implementation. -
DNADataReader.java: This file contains code to read DNA sequence data in from a file, such as
TestData.txtorDNAData.txt. - ValidatedInputReader.java: This file may be used for input of integers and strings.
- Debug.java: This file may be useful for testing and debugging your program.
IMPLEMENTATION
The implementation of this program has been divided into smaller tasks (stages), which should make testing easier.
Stage 1: Reading in the Data
Using an ordinary text editing application (such as Notepad, TextEdit, or even Word), look at the contents of the two data files provided above. Notice how they have a similar structure, but one is much simpler than the other. Make sure that you can recognize the different DNA sequences and their components (a GI identifier, a description, and then a sequence of nucleotides).
Create a class called TestDNAReader that you will use to test the early stages of this program as you develop it. This class should have a main method that asks the user for a filename, using the ValidatedInputReader class. It will then create a DNADataReader object with the file name entered by the user. It should then call the readData method, which for now simply prints some information about each sequence it reads in.
- Create a
mainthat just prints a simple string and test that. -
Use the
Debugclass for debugging output: turn on debugging (Debug.turnOn();)at the top of yourmainmethod, and then useDebug.printlnrather thanSystem.out.println. - Construct your
ValidatedInputReaderobject, ask the user for a filename, and test this by printing the filename. -
Construct a
DNADataReaderobject forthe appropriate filename, and test that the - Reminder: You may use the template for a main class and template for a normal class if you want, to get started.
readData method does what you expect. (Note that the initial version of DNADataReader will only produce debugging output if you have turned debugging on.)
Create a DNASequence class for DNA sequence objects. (What attributes should a DNA sequence object have? How will those attributes get initialized? What should the constructor do, and what parameters does it need?) This class should allow the user to access the sequence information, but users should not be allowed to modify it. (So, what methods should the class have?) The DNASequence class should also have a toString method that will return all of the sequence information (GI, description and nucleotide sequence) as a nicely formatted string.
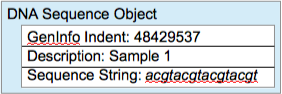
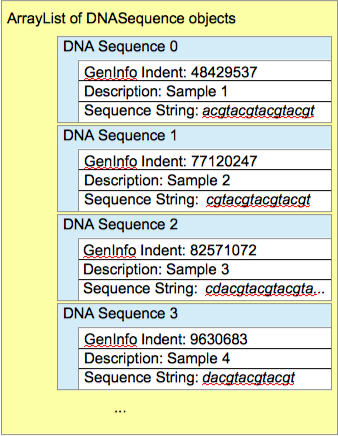
Look through the DNADataReader class. This class will be used to read the data from the two text files, but it is incomplete. Read the complete class to understand the nature of the changes you will be making. You will probably then find it easiest to make the changes "bottom-up": make the suggested changes in the lowest-level method first, then edit the method that calls it to correspond to the changes you just made, etc.. (A figure illustrating a list of DNASequence objects, such as you are asked to create in DNADataReader, can be found in Figure 3 below.)
Edit the TestDNAReader class so that it saves what is returned from the readData method into an ArrayList. Finally, it should iterate through the ArrayList and print out the data for each DNA sequence. Compare your output with the data file you used. If your output agrees with the file, you're ready to continue.
Submit your code in a zip file on Kit before moving on to Stage 2.
Stage 2: Create additional classes
The overall goal of this project is to report on the locations where various substrings can be found in a set of DNA sequences. To keep track of locations, create a Location class that will hold a pair of integers indicating (a) the DNA sequence string, and (b) the starting location of the substring within the sequence string. For instance, in the example at the top of this lab, the substring acgt occurs at locations 0, 4, 8, and 12 in the first DNA Sequence string (string 0) and at locations 3, 7, and 11 in the second (string 1). The Location objects representing these locations would consist of the following (string, location) pairs: (0, 0), (0, 4), (0, 8), (0, 12), (1, 3), (1, 7), and (1, 11).
Assume that Location objects will be created only after both the sequence number and starting location are known. What should the constructor look like, and what parameters does it need? Assume also that the attributes of Location objects are not changed after the object is created. What does that tell you about what methods are needed or not needed? In addition, you should create a toString method that returns the contents of a string in the (string, location) format used above, e.g., (0, 12).
To keep track of the locations associated with a number of different substrings, it will be useful to have a class that bundles a particular substring along with the list of its locations.
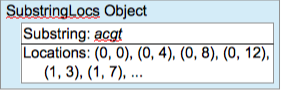
Give this class an appropriate name, such as SubstringLocs. What attributes should this class have? Assume that objects of your new class will be created with the substring value specified but before the locations have been determined. (Alternative: A different, valid assumption would be that each new object is created with a substring and a single, initial location for the list.) What should the constructor do, and what parameters does it need? Should this class have methods that let client code change the substring? Should it have methods that let client code add, remove, or change locations in the list of locations? What about a toString method? One simple toString format for the object in Figure 2 would be
acgt: (0, 0) (0, 4) (0, 8) (0, 12) (1, 3) (1, 7)
Be sure you have included adequate and appropriate documentation for all your classes.
Testing: Add code to your main method that tests your Location and SubstringLocs classes as you develop them. You can test toString() methods by calling them explicitly or by using the objects in System.out.print statements. For example,
System.out.println(new Location(3, 4));
tests both the Location constructor and its toString method.
Submit your code in a zip file on Kit before moving on to Stage 3.
Stage 3: Indexing
Create an Indexer class that will be used to find all of the n-letter substrings and their locations within a set of DNA sequences.
The constructor of the Indexer class should do the following:
- Ask the user for the name of the data file.
- Ask the user what length substrings to use.
- Create a
DNADataReaderobject with the user-specified information. - Obtain an
ArrayListcontaining the DNA sequences. (Where will you get it? From which object?) - Construct an empty
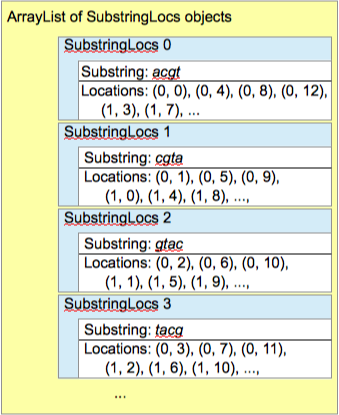
ArrayListthat will hold theSubstringAndLocsobjects. - Figures illustrating the two lists are below.
Reminder: Think about where the references to these objects should be declared. Are they local variables, or should some or all of them be instance variables? Remember what the task of a constructor is.
Add a run method to the Indexer class. This method should scan through the nucleotide sequences, looking at all the different substrings of the length specified by the user and building up a list of substrings and their locations, as illustrated in Figure 4 below. When a given substring is encountered for the first time, it should be added to the list along with its location. If the substring was already in the list, its location should be added to the appropriate location list. (You may find it helpful to create a private helper method that, for a given substring, finds the associated substring-and-locations object in the list. It would also, of course, indicate when the given substring is not yet represented in the list.)
Add a printResults method to the Indexer class. The method should print a list of each substring found, along with the locations at which it appears.
Finally, create a test class and name it Project1Tester, with a main method that creates an Indexer object, tells it to run and prints the results. Test your program (if using BlueJ, click on the main method of the Project1Tester class). If you use the TestData.txt file and substrings of length 4, you should see and output similar to this (depending on how you wrote the toString method of the SubstringLocs class):
acgt: (0, 0) (1, 0)
cgtg: (0, 1) (1, 1) (2, 0)
gtgc: (0, 2) (1, 2)
tgct: (0, 3) (1, 3)
gctg: (0, 4) (1, 4) (2, 31)
ctga: (0, 5) (1, 5)
tgac: (0, 6) (1, 6)
gaca: (0, 7) (1, 7) (2, 7)
acag: (0, 8) (1, 8) (2, 8)
cagt: (0, 9) (1, 35) (2, 19)
agtc: (0, 10) (2, 20)
gtcg: (0, 11) (0, 23) (1, 12) (1, 23) (2, 12) (2, 21)
tcgt: (0, 12) (0, 24) (1, 13) (1, 24) (2, 13) (2, 22)
cgta: (0, 13) (0, 25) (1, 14) (1, 25) (2, 14) (2, 23)
gtag: (0, 14) (2, 15) (2, 24)
tagt: (0, 15) (2, 25)
agtt: (0, 16) (0, 31) (1, 31) (2, 26)
gtta: (0, 17) (2, 27)
ttaa: (0, 18)
taag: (0, 19)
aagg: (0, 20) (1, 20)
aggt: (0, 21) (1, 10) (1, 21) (2, 10)
... and so on.
Stage 4: Submission
When you are finished, create a zip file from your project and submit the zip file on Kit. Name this zip file project1_yourname.zip. You should include in this zip file a copy of the output obtained when running your program with the DNAData.txt file, using substrings of length 7. You may choose to print only the substrings that occur more than once. Please copy the output after your program runs and then save it in a file that you will name output.txt.
EVALUATION
Project #1 is worth 30 points. The following items describe the criteria we will use to evaluate your project:
- Stage 1 (5 points):
- Correct use of
DNADataReaderclass 2.5 pts. - Correct implementation of
DNASequenceclass 2.5 pts.
- Correct use of
- Stage 2 (5 points):
- Correct implementation of
Locationclass 2.5 pts. - Correct implementation of
SubstringLocsclass 2.5 pts.
- Correct implementation of
- Stage 3 (10 points):
Indexerclass: correctly implementing theconstructorfor this class 4 pts.Indexerclass: correctly implementing therunmethod 4 pts.Indexerclass: correctly implementing theprintResultsmethod 2 pts.
- Stage 4 (10 points):
- The output file should contain the correct answer for the
DNAData.txtfile using substrings of length 7 8 pts. - Main application (containing the
mainmethod and the starting point) 2 pts.
- The output file should contain the correct answer for the
- Note: Make sure these classes are properly implemented, you have clean code, and the appropriate amount of comments.
Submit your completed program through Kit, under Project 1: Indexing Substrings.
Have fun! And if you have any questions remember that you can ask questions via email, Teams, and/or the Collaboration Center!
← back to the schedule